Project Z02
Studying in situ tissue schematics using advanced volumetric imaging and machine learning for image analysis
Project Description
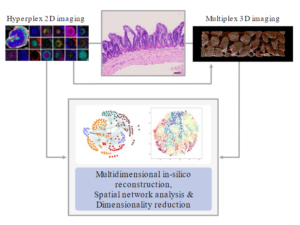
Inflammation begins in the tissue, and its shape and extent are largely determined by it (Farber, Nature 2021; Matzinger & Kamala, Nat Rev Immunol 2011). Infiltrated, migratory immune cells are highly plastic and dynamically integrate molecular and biophysical information from their environment that ultimately determines their effector functions and downstream fate. With the advancement of high-dimensional OMICs approaches, we started to appreciate the cellular heterogeneity of complex inflammatory infiltrates. While recent computational approaches can predict cellular interactions based on transcriptomic profiles, their actual functional neighborhoods and imprinting microanatomical environments remain speculative. To understand the relationship between tissue structure, microenvironment architecture, and cell function, multidimensional cell profiling must be complemented by quantitative in situ imaging to incorporate actual spatial biology and tissue organization. Here, current 2D hyperplex imaging techniques enable the acquisition of multiple parameters with spatial context. However, these techniques do not capture the complex 3D architecture of structurally multilayered and compartmentalized tissues, such as the gut or brain. At the same time, the wealth of data generated by these imaging and OMICs approaches entails subsequent analysis steps with nearly prohibitive efforts when aiming for manual assessment. To bridge these gaps, we seek to establish new imaging techniques and quantitative, machine-learning supported analysis approaches for the tissues across the gut-brain axis with high granularity in both 2D and 3D. This exploratory work will help integrate spatial data with cellular profiling data from this consortium to decipher the underlying tissue schemas in health and disease (Bhate et al., Cell Syst 2022).
Project Leaders:
Post Doctoral Researcher: